
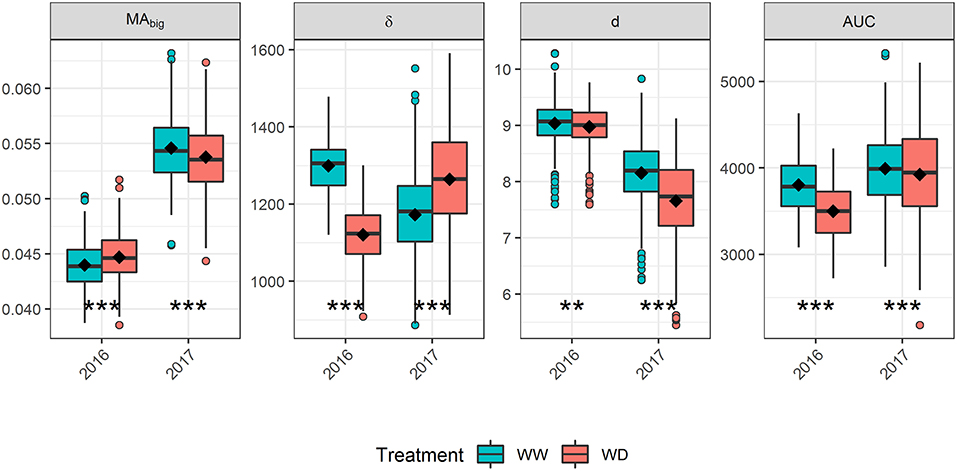
May associate singularities in the A matrix with random effects Is designed to help as without it, reordering The issue is to get the degrees of freedom correctĪnd to get the correct calculation of the Likelihood, especially inīivariate cases where DF associated with groups may differ between traits. Where the supergroup fixed effect is formally fitted separately in the model.Ī constraint is added to the inverse whichĬauses the preceding set of groups which have members to have effects You may insert Groups with no members to defineĬonstraints on groups, that is to associate groups into supergroups The first g lines of the pedigree identify genetic groups (with zero in both the sire and dam fields).Īll other lines must specify one of the genetic groups as sire or dam if the actual parent is unknown. Use !Goffset -1 to add no offset but to suppress insertion of constraints Typically, o is small as it represents genetic variance relative to group variance. When a constant is added, no adjustment of the degrees of freedom is made for To the diagonal elements of A inverse pertaining to groups. (see !GROUP below) is to shrink the group effects by Instructs ASReml to write out the A-inverse in the format of Where 0 ltf lt1 is the inbreeding level of such individuals. Individuals unless their inbreeding level is set with !FGEN f The first field of the pedigree file), they will be assumed base non-inbred Has implicit individuals (they appear as parents but not in Individual is taken as its inbreeding value. Or the level of inbreeding in a base individual.Ġ indicates a simple cross, 1 indicates selfed once,Ģ indicates selfed twice, etc. Indicates the individuals in the pedigree are inbred to some degree.Ĭontains a fourth field indicating the level of selfing Identities are alphanumeric with up to 120 characters otherwise by default they are numeric whole numbers <200,000,000.ĭiagonal elements of the Inverse of the Relationship Matrixįor the individuals (calculated as the diagonal of A-I)
FIXED EFFECTS ASREML STRATUM FREE
is read free format it may be the sameįile as the data file if the data file is free format and has the.Pedigree of an individual appears before any line where that The individual is required if the !XLINK qualifier is specified, Used if the !FGEN qualifier is specified, an optional fourth field may supply inbreeding/selfing information.Individual, its sire and its dam (or maternal grand Qualifier is associated with a data field. Relationships for fitting a genetic animal model and is required if The pedigree file is used to define the genetic Typically additional individuals providing additional genetic links are present ) appears as the first three fields of the data file, the dataįile can double as the pedigree file. Together with the corresponding lines of the data fileĪll individuals appearing in the data file must Is specified after all field definitions and before That the corresponding data field has an associated pedigree. In this chapter we consider data presented in Harvey (1977) using Provide a particular general relationship matrix (GRM) or its incverse (GIV) explicitly in a Relationship matrix is not the appropriate/required matrix, the user can Users new to this subject might find notes by Julius van der Werf helpful:įor the more general situation where the pedigree based inverse Matrix that is formed by ASReml for analysis. The additive genetic relationship matrix (sometimesĬalled the numerator relationship matrix) can be calculated from Provided all the genetic links are in the Genetic effects can be derived from the pedigree Modes of inheritance, the correlation expected from additive Genetic effects are therefore correlated and, assuming normal Set of animals that are genetically linked via a pedigree. `sire model' genetic analysis we have data on a


 0 kommentar(er)
0 kommentar(er)
